Analytical Protein Biochemistry: Robert Shoeman
The performance of instrumentation for the precise biochemical characterization of proteins has increased dramitically in recent years, primarily as a result of improvements in computer hardware. This is particularly obvious in the field of mass spectrometry, where newer instruments show increases in resolution and sensitivty of more than 1000-fold over instruments dating from the year 2000, and in robotics, where sophisticated as well as dedicated instruments have become affordable and usable, even for small groups of users.
4DX XSpectra

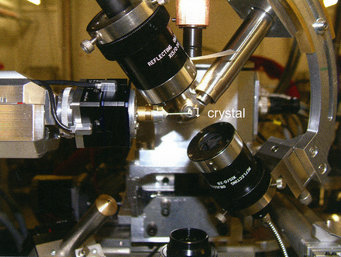
The XSPECTRA microspectrophotometer installed at the protein crystallography X10SA beamline at the swiss light source, Villigen, Switzerland. The optical axis and the synchrotron beam axis intersect within a 20 µm spot on which the crystal is centered.
In other experiments (in collaboration with Ilme Schlichting and Jochen Reinstein Group), we have combined micro optical spectroscopy with X-ray diffraction analysis and mass spectroscopy. Our department's microspectrophotometer is a further development (originally by Georg Holtermann, Max Planck Institute of Molecular Physiology Dortmund) of the commercial 4DX XSPECTRA system and can be used in a stand-alone mode or at the synchrotron beamline (Figure 4).
This makes possible a variety of sequential measurements, two of which are shown in Figure 5. The UV-VIS spectrum of a single protein crystal of a blue light photoreceptor, the BLUF protein BlrB, was measured at 100 K, confirming that the dark adapted ground state of the receptor had been crystallized (Figure 5, left side); thereafter, the crystal was transferred to a Maldi target, washed and analyzed. Unexpectedly, 2 peaks are seen with a difference in m/z values of 785 Da. This mass difference is that expected for a FAD cofactor. Further experiments have shown that the acidic matrix solution effectively extracts much of the bound FAD (it can be found in wash solutions). Irrespective of this experimental nuance, the result presented in Figure 5 clearly shows that FAD (m/z 785) and not FMN (m/z 458) is the cofactor bound to the protein in the crystal, which is important as there is no electron density for the terminal AMP part. This observation confirms the result of conventional HPLC assays that have been performed on this and other similar proteins, but which cannot be performed on small, single crystals.
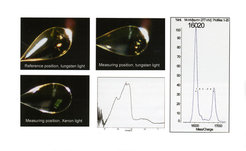
A crystal of a BLUF domain protein in various positions on the microspectrophotometer and its UV-VIS spectrum (left). Maldi TOF analysis of the same crystal (right) demonstrates that this protein contains bound FAD and not FMN.
Scientific Outlook
We recently supplemented our mass spectrometers with a Proteome Biosystems Xcise gel processing robot, which performs automated gel spot picking, peptide map fingerprinting (PMF) and spotting of purified peptides onto 394 position Maldi targets.
Current efforts also include the implementation of in-house computaional services (Mascot Server from Matrix Science) to permit fully automated PMF searches and authentication of protein, peptide and domain identities. Our sjort term plans include experiments with new matrices of sample substrates which function without matrix, to permit the simple and sensitive detection of low m/z compounds (such as cofactors and metabolites). Further efforts will include offering advanced mass spectrometry training courses for department and, if the demand is sufficient, institute members. Longer term plans include the use of complementary methods for protein characterization based on first principles, such as analytical ultracentrifugation.