Comparison of DnaK system regulation by different organisms
One key feature of the DnaK system is an ATP-driven cycle that alternates in its substrate binding properties between slow (holding) and fast exchange (release/folding).
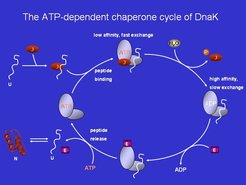
This cycle is regulated by GrpE/Hsc20 (nucleotide exchange factor) and DnaJ/Hsc40 (ATPase stimulating factor). We aim to understand the general features of this cycle, but also specific properties that result from adaptation of various organisms to their environmental needs and reflect the available repertoire of chaperones encoded by their genomes. The methods employed include functional assays (e.g. micro-titer plate analyses of ATPase activity or refolding with different substrate proteins), but also measurement of all key rate-constants using transient kinetic methods (e.g. stopped-flow, quenched-flow) and specific fluorescent labels which are attached to the components, such as the nucleotide, the substrate protein or peptide, or the chaperone.
The organisms investigated so far comprised E. coli, Thermus thermophilus and recently the chaperones of the archaebacterium Methanothermobacter thermautotrophicus ∆H that has a highly reduced set of chaperone systems. This ‘minimalist’ has to rely largely on a small heat shock protein (sHsp), a thermosome component, and the DnaK system; it is therefore an ideal system in which to study basic rules and interactions of chaperone mechanisms.
In the last years, we have custom-designed and synthesized fluorescently labeled nucleotide analogues and have used them to also address mechanistic questions regarding the Hsp90 chaperone system (in collaboration with Johannes Buchner, TU Munich). The nucleotide dependent balance and kinetics of interconversion of the functional states of Hsp90 are particular focus.

