Virus Capsid Assembly and Molecular Chaperones: Jochen Reinstein
Abstract: Virophages are small DNA viruses that replicate in unicellular eukaryotes that are co-infected with giant viruses. They recruit the cytosolic reproduction and assembly machinery of the giant virus whose own propagation is then stalled (see also Working group Matthias Fischer/BMM). Up to now 3 different virophages, namely Sputnik, Mavirus and Zamilon were isolated (1-3). They contain about 20 coding regions in a DNA genome of approx. 20 kb. The most conserved components are two capsid proteins, a protease that presumably assists maturation and a FtsK-HerA type ATPase that supposedly is responsible for ATP dependent DNA transport into the assembled capsid.
We characterize key features of Mavirus capsid assembly and processing by the respective protease in vitro and in vivo. In continuation with previous studies about protein folding and folding assistance through molecular chaperones, the folding and assembly/disassembly pathways of viral capsid proteins are characterized with spectroscopic (light scatter techniques, Fluorescence (also FRET (4)), CD) and structural methods (X-Ray, EM) with the aim to improve our knowledge on the hierarchy of supramolecular organisation of the virophage capsid. This also includes molecular insights into function and structure of the DNA packaging process which is supposed to be mediated by a FtsK-HerA type hexameric ATPase motor protein (similar to the AAA+ ClpB chaperone (5)) and processing of the capsid by the viral Cys-protease.
References:
[1] Zhang X, Sun S, Xiang Y, Wong J, Klose T, Raoult D, et al. Structure of Sputnik, a virophage, at 3.5-A resolution. Proc Natl Acad Sci U S A. 2012;109:18431-6.
[2] Fischer MG, Suttle CA. A virophage at the origin of large DNA transposons. Science. 2011;332:231-4.
[3] Gaia M, Benamar S, Boughalmi M, Pagnier I, Croce O, Colson P, et al. Zamilon, a Novel Virophage with Mimiviridae Host Specificity. Plos One. 2014;9.
[4] Muller B, Anders M, Reinstein J. In vitro analysis of human immunodeficiency virus particle dissociation: gag proteolytic processing influences dissociation kinetics. PLoS One. 2014;9:e99504.
[5] Zeymer C, Fischer S, Reinstein J. trans-Acting arginine residues in the AAA+ chaperone ClpB allosterically regulate the activity through inter- and intradomain communication. J Biol Chem. 2014;289:32965-76.
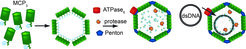
Fig 1: Schematic view of our working model for Mavirus Assembly, DNA-transport and Capsid processing (Figure provided from Diana Born).
Attaining a well defined three dimensional structure and thus functionality can be a serious challenge in the early life of many proteins. Although the final structure is energetically favored, many side reactions can occur mostly leading to aggregation that prevent the formation of the native protein structure.
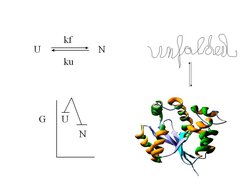
Molecular chaperones are ubiquitous in prokarytic/eukaryotic organisms and form cellular networks which assist protein folding in the cell. The underlying mechanisms, often dependent on the chemical energy of ATP, are diverse and complex. The DnaK/Hsc70 system on its own is mainly involved in preventing aggregation of challenged protein substrates, but in cooperation with ClpB/Hsp104 it can also rescue already aggregated proteins.
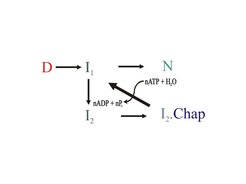
Protein Folding – chaperones help to prevent and revert unproductive pathways
The individual folding properties and potential recognition sequences of target proteins, raise the question about overall balance versus optimization of chaperon assisted folding.

![[""] © Jochen Reinstein](/16788/original-1314699626.jpg?t=eyJ3aWR0aCI6MzQxLCJmaWxlX2V4dGVuc2lvbiI6ImpwZyIsIm9ial9pZCI6MTY3ODh9--881709d3d6803ecc6ba1407dc960145bf5a12626)
![[""]
Protein Folding – chaperones help to prevent and revert unproductive pathways](/93737/original-1421399972.jpg?t=eyJ3aWR0aCI6MzQxLCJmaWxlX2V4dGVuc2lvbiI6ImpwZyIsIm9ial9pZCI6OTM3Mzd9--f64bc0fe44ef4e142e9a02ab9930c3390c25db43)