Protein De-Aggregation - Structure and Mechanism of the DnaK/ClpB System

Model for concerted mechanism of ClpB/DnaK mediated deaggregation
© Jochen Reinstein
The mechanism by which the DnaK/ClpB system can solubilize and renature aggregated proteins is a centrale theme in our group.
The functional role of ClpB in actively untangling aggregates in an ATP-dependent process implies that this chaperone must have the properties of a molecular motor. This means that ATP hydrolysis is coupled to mechanical movements which somehow alter the structural properties of protein aggregates.
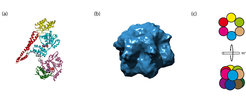
Structure and oligomeric assembly of hexameric AAA+ protein ClpB. (1QVR.pdb and EMD-1241))
© Jochen Reinstein
In order to improve our understanding of the ClpB mechanism, we are currently trying to obtain specific dyanmic and structural information on its different nucleotide states (static and dynamic light scattering, transient kinetic studies using fluorescent probes, FRET, anisotropy, XRay).

