Tracking molecular movements with nanometer precision using MINSTED
Researchers led by Stefan Hell at the Max Planck Institute (MPI) for Multidisciplinary Sciences in Göttingen and the MPI for Medical Research in Heidelberg have succeeded in using MINSTED microscopy to visualize movements of the motor protein kinesin with spatio-temporal precision in the nanometer per millisecond range. Hell’s team has thus extended the MINSTED imaging method to precise tracking of molecular movements.
It was like a revolution in light microscopy. With STED microscopy, Hell fundamentally broke the diffraction resolution limit of fluorescence microscopy previously considered impossible. For this breakthrough the physicist was awarded the Nobel Prize in Chemistry in 2014. In its original version, STED microscopy achieved a resolution of up to 20 nanometers – around ten times sharper than all light microscopes available at the time.
In 2016, Hell and his team succeeded to increase the resolution tenfold again: For the so-called MINFLUX nanoscopy, they combined the use of a fluorescence excitation beam shaped like a donut (as in STED microscopy) with the detection of single fluorescence molecules, and thus achieved a resolution of just a few nanometers for the first time. MINFLUX can truly make fluorescent molecules visible on molecular scales – it cannot get sharper.
With developing MINSTED, Hell and his co-workers presented another high-resolution microscopy method in 2021 that can separate fluorescently labeled details with molecular sharpness. Using a regularly focused beam for fluorescence excitation and a donut-shaped STED-beam for fluorescence suppression, MINSTED incorporates even more of the original STED principle than MINFLUX. This brings advantages. “Like MINFLUX, MINSTED achieves molecular resolution but is better at suppressing background. Thus, it provides a better signal-to-background ratio (SBR),” explains Henrik von der Emde, a PhD student in Stefan Hell's Department of NanoBiophotonics at the MPI for Multidisciplinary Sciences and one of the first authors of the study now published in the journal Nature Methods. “As it requires far fewer detected fluorescence photons, MINSTED also localizes the position of single fluorescent molecules much faster than established methods relying on detection with a camera.” So far, however, MINFLUX has been superior to MINSTED in one respect: tracking the dynamics of individual proteins in detail.
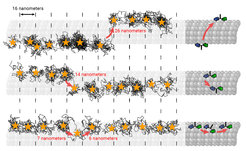
The scientists from Göttingen and Heidelberg have now demonstrated that MINSTED also has this potential using the small motor protein kinesin. In cooperation with other motor proteins, kinesin transports biological cargo in cells. The researchers have now succeeded, for the first time, to use the speed of MINSTED to “film” kinesin in action. This enabled them to visualize how the motor protein moves along the cell’s “transport roads” , the microtubules. They thereby achieved a similar spatial-temporal resolution in the nanometer/millisecond range as with MINFLUX. The main advantage of MINSTED, however, is its ability to suppress background. This improves the SBR by an order of magnitude over that achieved by MINFLUX. The improved SBR could make MINSTED more suitable for samples with an inherently higher background.
“The tracking is so powerful that we even observed rare cases of ‘lane switches’, where kinesin walking on one protofilament, which is a lane of the microtubulus, jumps sideways onto another and continues walking there,” says Lukas Scheiderer, former PhD student and now postdoctoral researcher in Stefan Hell’s Department of Optical Nanoscopy at the MPI for Medical Research and also first author of the study. This was made possible by recording the protein movements with a spatio-temporal precision of up to a few nanometers per millisecond. To identify such tiny changes in position, the microscope only needs to detect around 15-20 photons emitted by the fluorescent molecule.
“With MINSTED and MINFLUX, we now have two very powerful techniques at our disposal that enable the highest spatial and temporal precision currently available,” Hell says. Movements and changes in the structure of individual protein molecules can thus be investigated in a completely different way. No one method is generally superior to the other. Depending on the application, one or the other microscopy technique can play to its strengths. (cr)
(Press Release by the MPI for Multidisciplinary Sciences)
