
Equipment
My group is interested in the biophysical characterization of peptides and proteins (and their associated cofactors) and we work in close cooperation with all of the reaerch groups in our department; collaborations with other groups in the institute are starting.
Kratos matrix laser assisted Maldi TOF
Kratos matrix laser assisted desorption / ionization time-of-flight (Maldi TOF) Maldi IV mass spectrometer with a curved field reflectron and post-source decay (PSD) functions.
A standard application of the Maldi TOF mass spectrometers is the determination of m/z values for proteins during and after purification. An interesting example is seen in Figure 2 for recombinant Clp1, a protein involved in 3'-end processing of mRNA (Anton Meinhart Group). While the desired protein seemed quite pure in both SDS gel analysis and in the high m/z spectrum (Figure 2), further analysis showed the presence of significant amounts of peptides derived from the full-lenght protein. This information facilitated the choice of subsequent purification steps.
Maldi TOF analysis of a partially purified recombinant protein yielded the expected m/z value (left) and a number of lower m/z contaminants (right). Peptide ladder analysis generated the sequence shown (in one letter code), identifying the contaminants as breakdown products of the full-lenght protein.
Figure 2
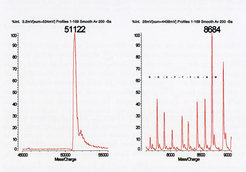
Maldi TOF analysis of a partially purified recombinant protein yielded the expected m/z value (left) and a number of lower m/z contaminants (right). Peptide ladder analysis generated the sequence shown (in one letter code), identifying the contaminants as breakdown products of the full-lenght protein.
Maldi TOF analysis of a partially purified recombinant protein yielded the expected m/z value (left) and a number of lower m/z contaminants (right). Peptide ladder analysis generated the sequence shown (in one letter code), identifying the contaminants as breakdown products of the full-lenght protein.
Improvements in mass spectrometer sensitivity have reduced the levels of detection for many peptides to the mid or high femtomole range and for many proteins to the low picomole or high femtomole range. This encouraged us to detect proteins present in single protein crystals, which helps to establish whether a crystal does indeed contain the protein or complex of interest before embarking on a lenghty optimization procedure or challenging X-ray data collection. Historically, such experiments have been hampered not only by the need to successfully manipulate the crystals but also by the presence of a variety of substances in the crystallization or cryogenic solutions (such as glycerol, polyethylene glycol or salt) which suppress the ionization of the crystal protein(s). We have developed several techniques for washing crystals and/or crystal proteins to permit their analysis via Maldi TOF.
Figure 3

Maldi TOF analysis of a partially purified recombinant protein yielded the expected m/z value (left) and a number of lower m/z contaminants (right). Peptide ladder analysis generated the sequence shown (in one letter code), identifying the contaminants as breakdown products of the full-lenght protein.
One example of such an analysis is Figure 3. Certain crystallization screen conditions result in the formation of a massive shower of rod-like crystals from a recombinant chaperone protein (Jochen Reinstein Group / Ilme Schlichting Group). Individual crystals can be teased out and washed (Figure 3B). After transfer to the Maldi plate, the crystal is dissolved and analyzed (Figure 3C). Anylyses of the starting protein solution, the wash solutions and the dissolved crystal show that, in this case, the protein molecules in the crystal have m/z values identical to that of the starting material. In those quite common situations where this is not the case, PMF mass spectrometry has proven indispensable in identifying the missing amino acids, which can be invaluable for (re)interpretation of electron density maps and/or for new cloning/protein experiments.
Maldi TOF analysis of a single protein crystal. A crystallization reaction of a recombinant molecular chaperone protein prduces a shower of crystals (A), from which a single crystal was isolated (B) and subsequently washed. Maldi TOF analysis demonstrated that the protein in the crystal was identical to that in the starting solution.

